T. dohrnii on crab collected from Bocas del Toro, Panama
New Insights into the Biology of Cell Transdifferentiation
The ultimate goal of regenerative research is to replace damaged cells in response to injuries and aging. Transdifferentiation (or cell reprogramming), a process through which a mature somatic cell transforms into a new type of mature somatic cell, can achieve this goal.
The mechanisms by which cells spontaneously (in vivo) leave a differentiated state to become a new lineage are poorly understood. This is due to the difficulties in inducing transdifferentiation in live model systems.
A new promising approach comes from the cnidarian Turritopsis dohrnii. Most animals reproduce, age, and die. T. dohrnii has escaped this fate. When faced with unfavorable circumstances, the jellyfish of T. dohrnii avoid death by reverting to a younger life cycle stage, the polyp. During the life cycle reversal, which covers a time span of about 48-72 hours, cell transdifferentiation occurs.
These characteristics make T. dohrnii a potential new system for in vivo research on the molecular mechanisms of cell stability and transdifferentiation.
Our project will produce genomic tools and assess the potential of T. dohrnii as a system for the study of in vivo cellular transdifferentiation.

Research Objectives
Produce a transcriptome assembly and annotation of T. dohrnii
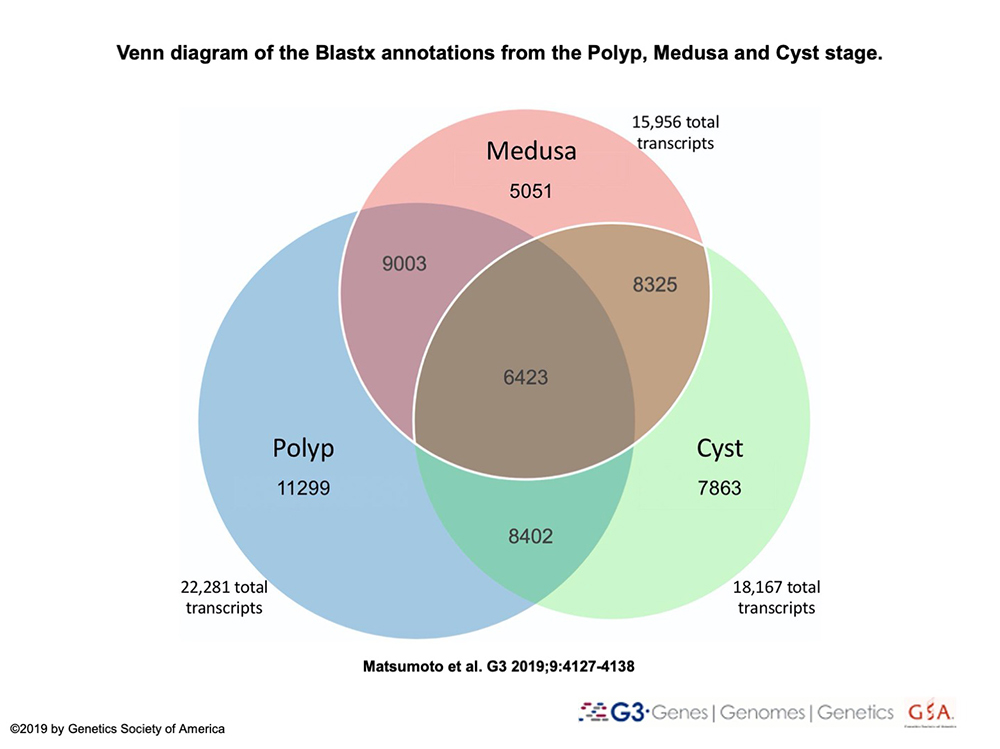
RNA-seq libraries of the colonial polyp, medusa, cyst and newly reversed polyp will be assembled into a transcriptome and characterized to further understand how gene expression changes throughout life cycle stages involved in reverse development.
Produce a hybrid draft genome assembly and annotation of T. dohrnii
Using the transcriptome assembly as a foundation, long DNA reads will be sequenced and combined to assemble a high-quality genome for T. dohrnii. The genome will be annotated and compared to other Cnidaria taxa to find species specific genes.
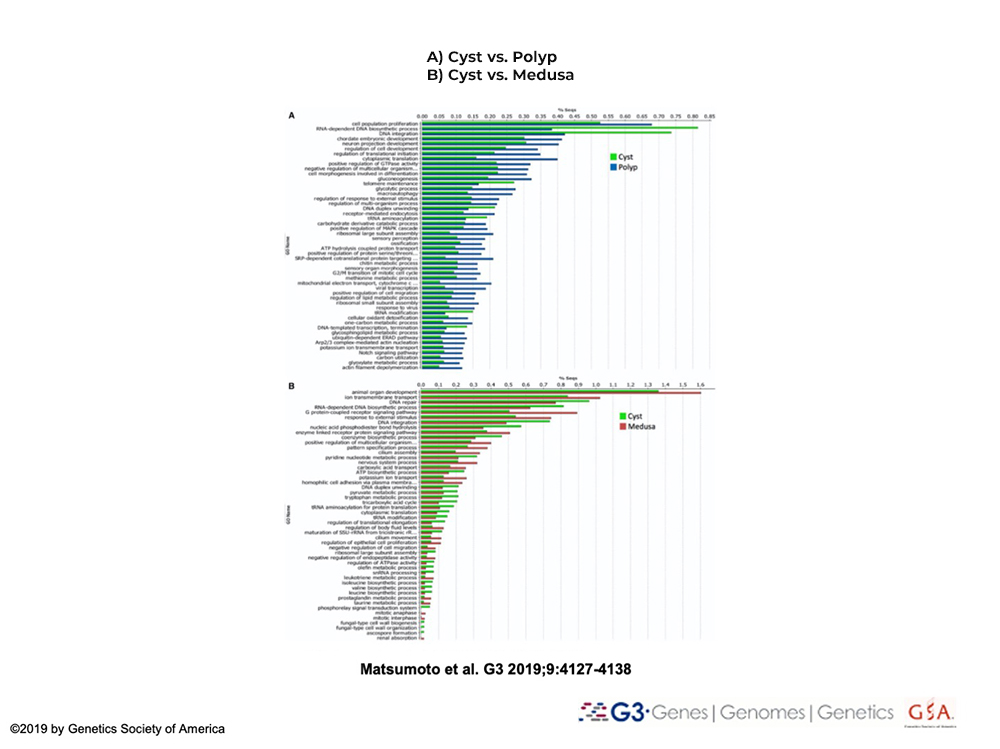
Conduct differential gene expression analyses of life cycle stages of T. dohrnii
Reverse development will be broken down in shorter intervals to identify genetic constituents and networks are involved in/or underlie control cell transdifferentiation and reverse metamorphosis in T. dohrnii.